|
Closing the gaps in the Streptococcus sanguis genome
Virtual PCR: Blast contig ends against a Streptococcus genome
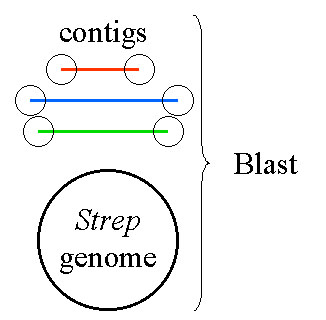
Your idea goes like this. If you extract the sequences from the ends of all 200 contigs and use them to Blast the genome of Streptococcus, you should be able to scan the hits for those that are nearby each other. This would identify ends that are close enough for PCR to work.
Of course the flaw in the plan is that it seems to require that you already know the very genomic sequence you're trying to determine!
Conceivably, however, the plan might work using the genomic sequence of a different strain of Streptococcus, one whose genome has already been determined, even though the genomes are similar to only a limited degree. You decide to give it a shot.
Back to main Scenario page back one page continue
|

