|
The origin of human genetic variability from
cross-species comparisons: the truth about Blast
What is a single nucleotide polymorphism?
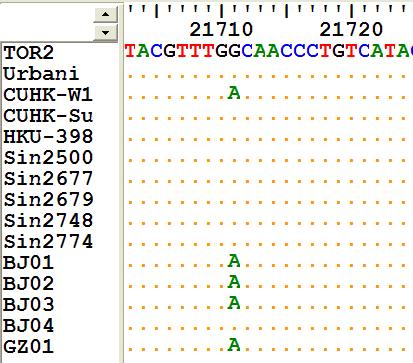 Single nucleotide polymorphism (SNP) is the result of single base change in a population (sample) of DNA sequences. SNPs are the most abundant genetic variants in the mammalian genomes. About 10 million SNPs are estimated in human populations. SNP data is also widely used in disease studies, population genetics, functional assays, genome mapping, mutational mechanisms, and genome evolution.
Single nucleotide polymorphism (SNP) is the result of single base change in a population (sample) of DNA sequences. SNPs are the most abundant genetic variants in the mammalian genomes. About 10 million SNPs are estimated in human populations. SNP data is also widely used in disease studies, population genetics, functional assays, genome mapping, mutational mechanisms, and genome evolution.
The physical location of the SNP in the genome or in the gene is important for genetic analysis. In comparative genomics, we may compare the mapped sites to determine the mutational directions using the SNPs that can be uniquely mapped in the different genomes. However, due to the nature of the genome sequence complexity, it may not be easy to map every SNP to the genome. In this research simulation, you will develop the procedures to map a set of SNPs to the human and chimpanzee genomes.
Back to main Scenario page continue
|

