FIGURE 1.
Sequence Specific Protein Binding to SRSR�s Present in Nostoc Punctiforme
Tom Murphy
VCU BBSI/Monmouth College
Introduction:
In the
past the majority of the focus of DNA research revolved around the DNA that
constitutes genes. However, with the advent of genomic sequencing much has
been learned about the sequences and functions of intergenic DNA as well.
Long thought of as "junk DNA" intergenic sequences have been found to exert
control over recombination, DNA replication, and gene expression (Van Belkum and
Scherer, 1998). Many repeats act as binding sites for proteins or as structural
elements on the level of RNA. Much of the intergenic DNA which regulates gene
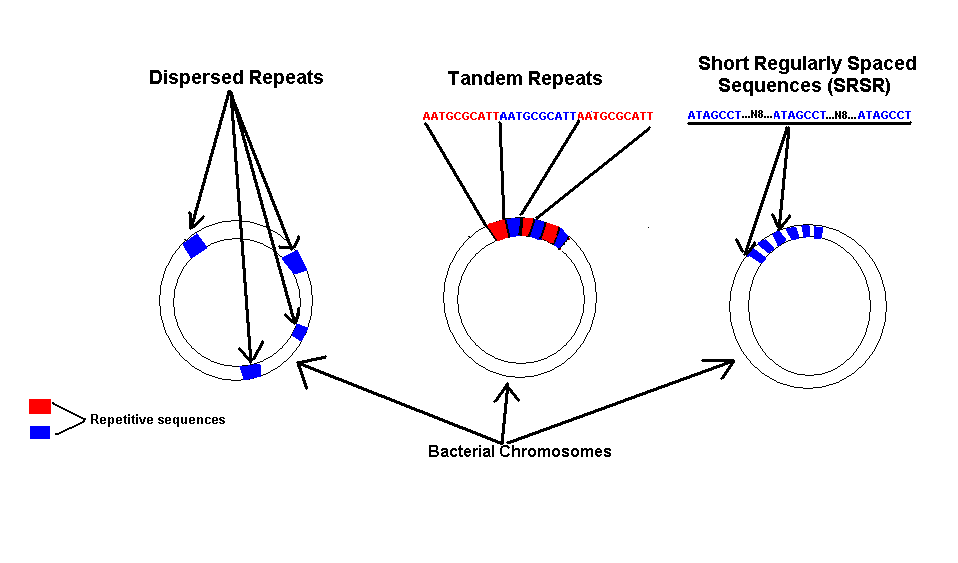
expression is found in the form of repetitive DNA. Repeats are classified
on the type of repeat (Fig 1) (dispersed, tandem,), the size of the repeat (long
or short), and their location and occurrence in the genome (Bachellier et al,
1999).
FIGURE 1.

While a significant amount of information is known about dispersed and tandem repeats little is known about other types of repetitive DNA. A new type of repeat has been characterized in many bacterial genomes but has been most studied in the archael genome of Sulfolobus solfataricus (Peng et al, 2003). The repeats found in these organisms show characteristics of both tandem and dispersed repeats. These repeats consist of short regularly spaced repeats (SRSR�s) of 20 to 37 bp separated by nonconserved sequences of almost constant length (inter-repetitive sequence) (Peng et al, 2003). The Sulfolobus repeat was found to be a binding site for an uncharacterized SRSR binding protein (Peng et al, 2003). Peng et al was able to determine that a protein binds to that specific SRSR found in Sulfolobus, however, they still know little about the function of the SRSR or the protein that binds to it.
Similar repeats have been found in the genomes of cyanobacteria Anabeana 7120 (Mesopohl et al, 1996) and Nostoc punctiforme. These repeats were found by accident while searching for dispersed repeats (Elhai, unpublished). This study will examine whether the repeats found in the cyanobacterium Nostoc punctiforme are similar to the repeats found in Sulfolobus solfataricus in the respect that they bind protein. Not only will the presence of bound protein be examined but also the specificity with which the protein binds to the SRSR.
Progress Report:
Over the past 10 weeks my goal was to examine many cyanobacterial genomes for SRSR�s and to further look at the repeats already found in Nostoc. The first question was how to locate these repeats. Many programs exist that enable you to search a genome for a specific nucleotide sequence but none existed that would enable me to look for SRSR�s without first knowing their sequence. My first step was to locate sequences that are similar to the original SRSR that I had. I had to determine how many mismatches I would accept and still consider the sequence to be similar. With the help of a program supplied by Jeff Elhai I searched the Nostoc genome for similar repeats allowing for up to 8 mismatches in the sequence. The results from this search were similar the same as I already had. The next step was to write a program that would allow me to search the genome for repeats based on their pattern (repeat, junk, repeat, junk�) rather than on their sequence. With the help of Jeff, I wrote such a program. The output from this program allows me to find all of the SRSR�s in a genome I have searched all available cyanobacterial genomes for SRSR�s.
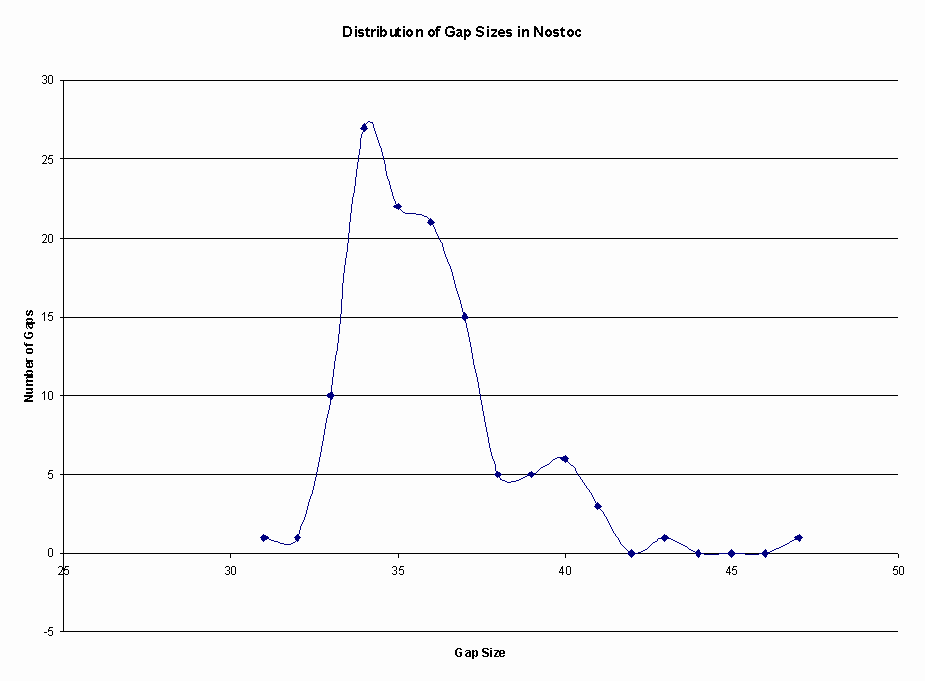
My findings show that this type of repeat is found in most cyanobacterial genomes and to some extent genomes bacterial genomes as well (Chart 1). Anabaena 7120, Nostoc Punctiforme, and Trichodesmium have the largest genomes so it is of no surprise that they contain the highest number of clusters as well. For this study a cluster is considered any repeat that contains at least 3 iterations of the repeat and 2 inter-repetitive regions. Analysis of the 3 most common repeats in Nostoc shows that the average inter-repeat region is around 36 bp (Figure 2).
Chart 1.
| Organism | Number of clusters* | Average Repeat size | average Gap Size (bp) | |
| Cyanobacteria | ||||
| Nostoc Punctiforme |
98 |
27 |
32 |
|
| Anabaena 7120 |
42 |
28 |
35 |
|
| Trichodesmium |
115 |
27 |
38 |
|
| ProchlorococcusMED |
5 |
27 |
29 |
|
| ProchlorococcusMIT |
7 |
27 |
26 |
|
| SynechocystisPCC6803 |
3 |
28 |
26 |
|
| Bacteria | ||||
| Bacillus |
2 |
20 |
41 |
|
| Ecoli K12 |
5 |
31 |
37 |
|
| *Cluster is three iterations of a repeat or more | ||||
Goals for Academic Year:
The overall goal of my research for the coming year is to determine if the SRSR found in Nostoc act as a protein binding site in hopes of better understanding the purpose of such repeats. By showing that they act as a binding site for proteins I would be able to draw a correlation between this SRSR and the one published for Sulfolobus. On the other hand if I show that no protein binds to the SRSR sequence in Nostoc I will demonstrate that SRSR�s may have different functions in different organisms. Either result will lead to more research next summer to better determine the function of SRSR�s. This could encompass either further characterization of the protein or a completely different approach to understanding SRSR�s. Further study will go into interpreting the data I have accumulated from searching the genomes for SRSR�s.
Plan for Academic Year:
Experiments
The first step in determining whether or not the SRSR found in Nostoc functions as a protein binding site is to perform a band-shift assay. I will follow a similar procedure to that described in Peng et al. I will PCR a section of the SRSR that contains 2 repeats and flanking regions. I will have to order PCR primers to extract such a segment. The 5� ends will be labeled using digoxen. A control lane containing a random unlabeled DNA sequence will be run to determine that if a protein binds it binds specifically to the SRSR and not just any DNA present. The DNA will then be reconstituted with purified cell extracts to allow any cellular proteins to bind to the DNA sequence. Two of the lanes will also contain unlabeled SRSR sequences. The DNA protein mixture will then be run on a 7% ployacrylamide gel and photographed. If a lighter first band is noticed in the lanes that contain unlabeled SRSR sequences it will show that protein is binding to the SRSR sequences. If a dark band is noticed in the control lanes this shows that the protein is binding specifically to the SRSR sequence. If a protein is present on the gel it will be purified for use in the footprinting assay.
The second experiment will involve footprinting the SRSR repeat to confirm that the protein binds to the SRSR repeat and not the inter-repetitive regions. I will again follow a procedure similar to that published by Peng et al. The SRSR fragment that was obtained by PCR and end labeled with digoxen will be reconstituted with the purified binding protein obtained from the band-shift assay. This mixture will then be digested with a nuclease (most likely DNase I) and run on a 10% polyacrylamide gel next to a sequencing ladder of the SRSR. The results will then be photographed and examined to determine if the protein binds over a part of the sequence in the SRSR.
Computation
An ongoing part of my research will include continuing to search available genomes for SRSR�s. Once all of the cyanobacterial genomes have been searched I will begin to examine other bacterial genomes. I will keep a record of all repeats found for later study.
Budget:
My budget includes all materials necessary for footprinting analysis and for band-shift assay. This rough estimate of this is $800 and my home institution will pick up the remaining costs.
References:
Bachellier, S., J. M. Clement, et al. (1999). "Short palindromic repetitive DNA elements in enterobacteria: a survey." Research in Microbiology 150(9-10): 627-639.
Mesepohl, B., Gorlitz, K., Bohme, H. (1996) "Long tandemly repeated (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. 7120." Biochimica et Biophysica Acta 1307: 26-30.
Peng, X., Brugger, K., Shen, B., Chen, L., She, Q., Garret, R.A. (2003) "Genus specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes". Journal of Bacteriology 185(8): 2410- 2417.
van Belkum, A., S. Scherer, et al. (1998). "Short-sequence DNA repeats in prokaryotic genomes." Microbiology and Molecular Biology Reviews 62(2): 275.