Development of a natural
language to connect humans to genomic data
With the acquisition of billions of nucleotides of DNA sequence information, We desperately need a language that will enable the vast majority of
researchers who do not program computers to make use of powerful tools
to analyze that genomic data. It is fanciful to expect that they will learn a conventional programming language, like C or Perl. They have not, despite great incentives to do so. I am part of a group that has developed a general purpose programming language, BioBIKE, that speaks in a way molecular biologists can understand and puts in their hands the tools of bioinformatics and the data of genomics. BioBIKE uses a graphical interface that takes advantage of the conventions familiar to even computer-averse biologists.
BBSI
project: Possible role of repeated sequences in the rapid
evolution of genomes
The genomes of closely related
organisms generally can be aligned with one another with few
distortions. Not so with cyanobacteria of the genus Nostoc! The
figure below shows a diagram of the completely sequenced genome of one Nostoc
(PCC 7120) aligned with a portion of the incompletely sequenced genome
of another Nostoc (N. punctiforme).
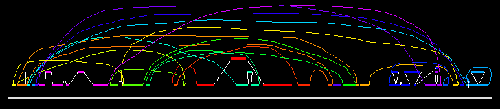
PCC 7120 genome
Alignment of PCC 7120 genome with 200 Kb of N.
punctiforme genome (gray)
It looks like the pieces of the genome have been put into an
evolutionary blender. How did this come about in so short a span of
evolutionary time? It turns out that Nostoc genomes are loaded
with repeated sequences and transposons (hopping DNA). Whether these
are the source of the evident genomic instability remains to be
demonstrated.
We can go back in history and determine the source of genomic
instability much in the same way an archaeologist makes inferences
about the human past. The truth of the matter is attainable by careful
attention to the molecular shards -- broken genes -- that litter all
genomes. These are the residue of past duplications and transpositions,
and from them, it is often possible to piece together what a genome
must have looked like in evolutionary time and thus deduce the forces
that continue to shape DNA sequences.
Other research interests
(see web page for more
details)
Mechanism of heterocyst spacing
in cyanobacteria
Toward a complete molecular and genetic understanding of what is
arguably nature's simplest example of multicellular pattern formation.
Signaling between plants and
symbiotic cyanobacteria
Some plants have learned how to domesticate N2-fixing cyanobacteria,
gaining thereby a ready nitrogen source. If we learn how these plants
do it, we might be able to pass the knowledge on to important crop
plants that otherwise rely on environmentally detrimental nitrogenous
fertilizer.


