|
BBSI project: In silico design of Erm inhibitors to thwart common antibiotic resistance?
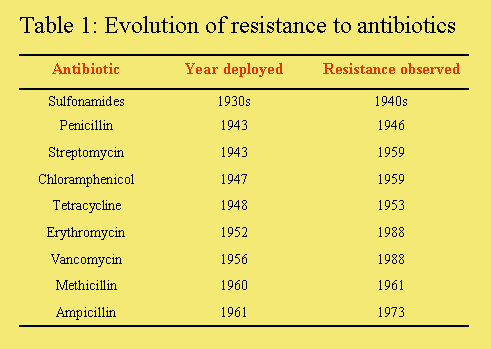
Antibiotic treatment of bacterial infections has benefited human health greater than any other drug therapy. Unfortunately, those gains have been put in jeopardy with the emergence of 'super-bugs', bacterial pathogens that are resistant to all or most of our current antibiotics. The development and passage of resistance, usually as a plasmid, from one organism to another is an improbable event, which nevertheless occurs as a result of the continual selective pressure exerted by antibiotic therapies. Basically, if a given antibiotic is used often enough for a long enough time, resistance will emerge against that antibiotic in one organism and spread to other organisms (Table 1).
There are several ways that rely on drug design to combat existing resistance and the spread or emergence of new resistance:
- Design or identify new therapeutics against novel targets. (
- Design modified or novel therapeutics against existing targets that somehow circumvent current resistance mechanisms.
- Design or identify agents that will inactive resistance mechanisms, thus allowing the use of existing drugs.
Choices 1 and 2 are the most commonly used strategies in the struggle against resistance, but choice 3 is also a worthwhile strategy and forms the basis for one of the research projects within my research group.
Bacteria carrying the protein Erm are resistant to erythromycin and several other medically important antibiotics. A drug that inhibits Erm could restore the usefulness of these antibiotics when co-administered. No anti-Erm drug currently exists, but the recent crystal structure of this enzyme allows for the computer-aided design of candidates that can be made and tested. A goal of our lab is to design a therapeutically useful Erm inhibitor.
Our project contains an important caveat. The inhibitor that we design must be so selective that it doesn't also inhibit an essential human enzyme, Dim1, that is remarkably similar in structure and function to the Erm protein, both acting to chemically modify RNA. We have recently solved the 3-D structure of Dim1 from another organism, so we are now in a position to design Erm inhibitors with full knowledge of how they might also inhibit Dim1. Therefore, selectivity can be designed in from the start. This is a challenging problem that will push the limits of in silico (computer) drug design, which, of course, makes it all the more exciting to work on.
Reference
Cashman DJ, Rife JP, Kellogg GE (2001). Which aminoglycoside ring is most important for binding? a hydropathic analysis of gentamicin, paromomycin, and analogues. Bioorganic & Medicinal Chemistry Letters 11:119-122.
close window |


